博文
2022 mLife 以微生物组工程原则为指导:环境利用的成就和展望
||
摘要
尽管微生物组工程的成就凸显了其对微生物群落的靶向操作的重要性,但知识和技术差距仍然限制了微生物组工程在生物技术中的应用,特别是在环境方面。应对难降解污染物和环境条件波动的环境挑战,需要充分了解微生物组工程的理论成果和实际应用。本文综述了近年来关于微生物组工程策略及其在生物修复中的经典应用的前沿研究。此外,还总结了一个框架,用于将微生物组工程中自上而下和自下而上的方法结合起来,以改进应用。建议设计用于环境用途的微生物组的策略,以避免对人类健康构成风险的有毒中间体的积聚。我们预计,突出的框架和战略将有利于工程微生物组,以应对困难的环境挑战,例如降解多种难降解污染物,并在波动条件下与本地微生物一起维持工程微生物组的原位性能。

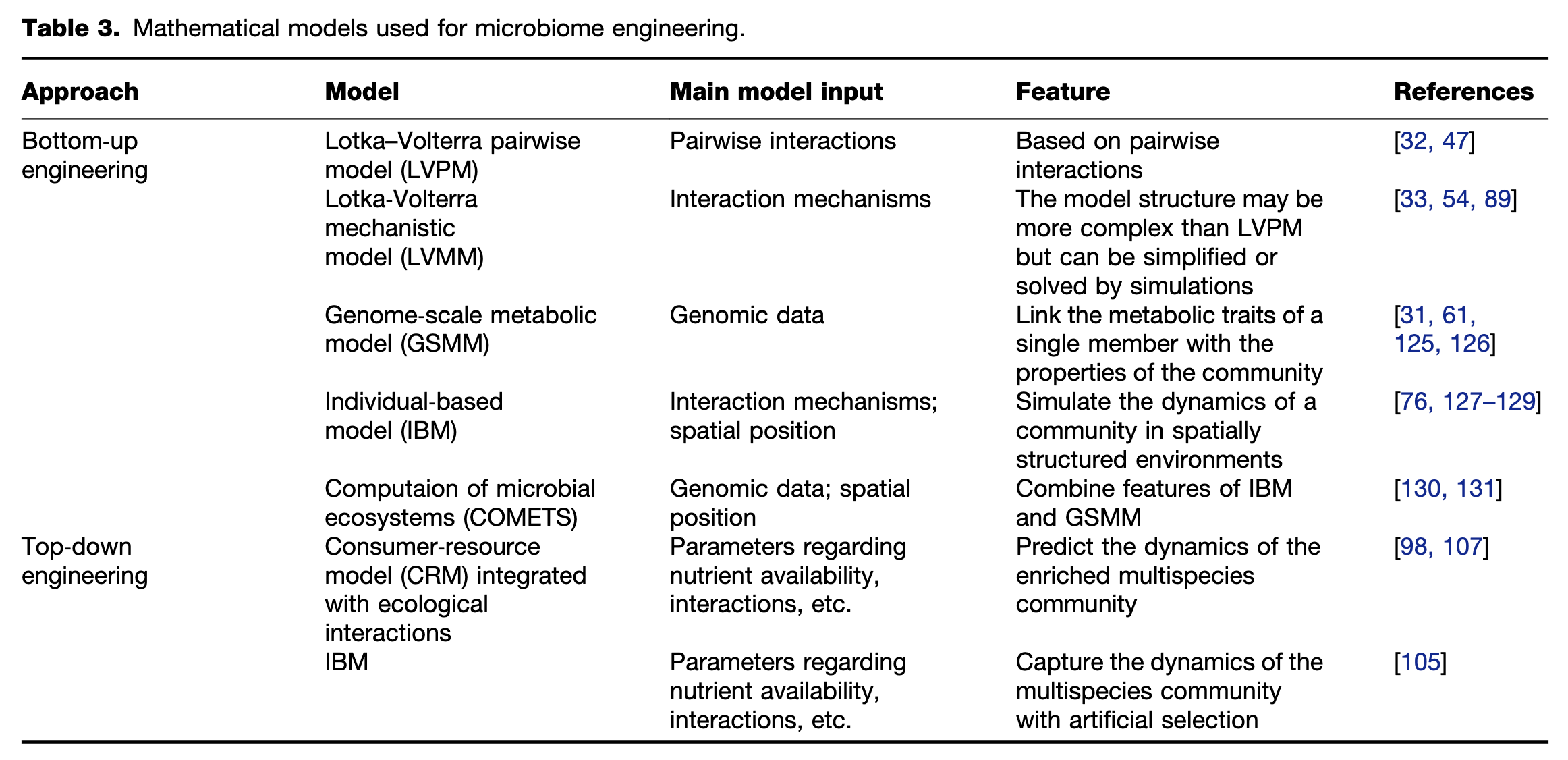
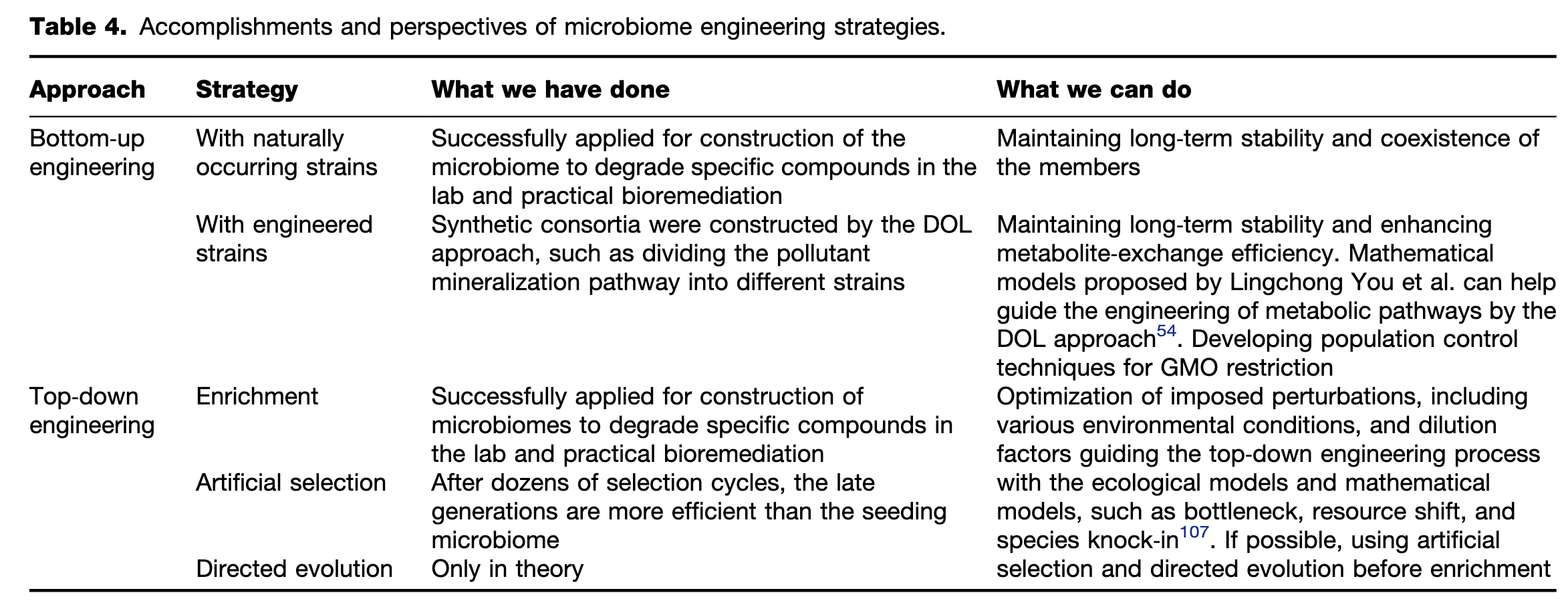
Figure 1 Two approaches to microbiome engineering. Left panel: the bottom-up approach of microbiome engineering starts with several isolates, whose physiologic features have been well characterized. Coculture assays are performed to identify pairwise interactions among these isolates. Genetic engineering could be performed to design or modify the interactions. Systems based on cell-to-cell communications are used to directly control the behavior of specific populations. Right panel: the top-down approach of microbiome engineering starts with a seeding microbiome containing uncultivated microorganisms. Then, microbiome engineering is managed to drive the seeding microbiome to self-assemble into a stable system with highly optimized function. Three methods, including enrichment, artificial selection, and directed evolution, are commonly applied to perform top-down engineering. No matter which approaches are used, the workflows should follow the “design-build-test-learn” (DBTL) cycle proposed by Lawson et al.13.
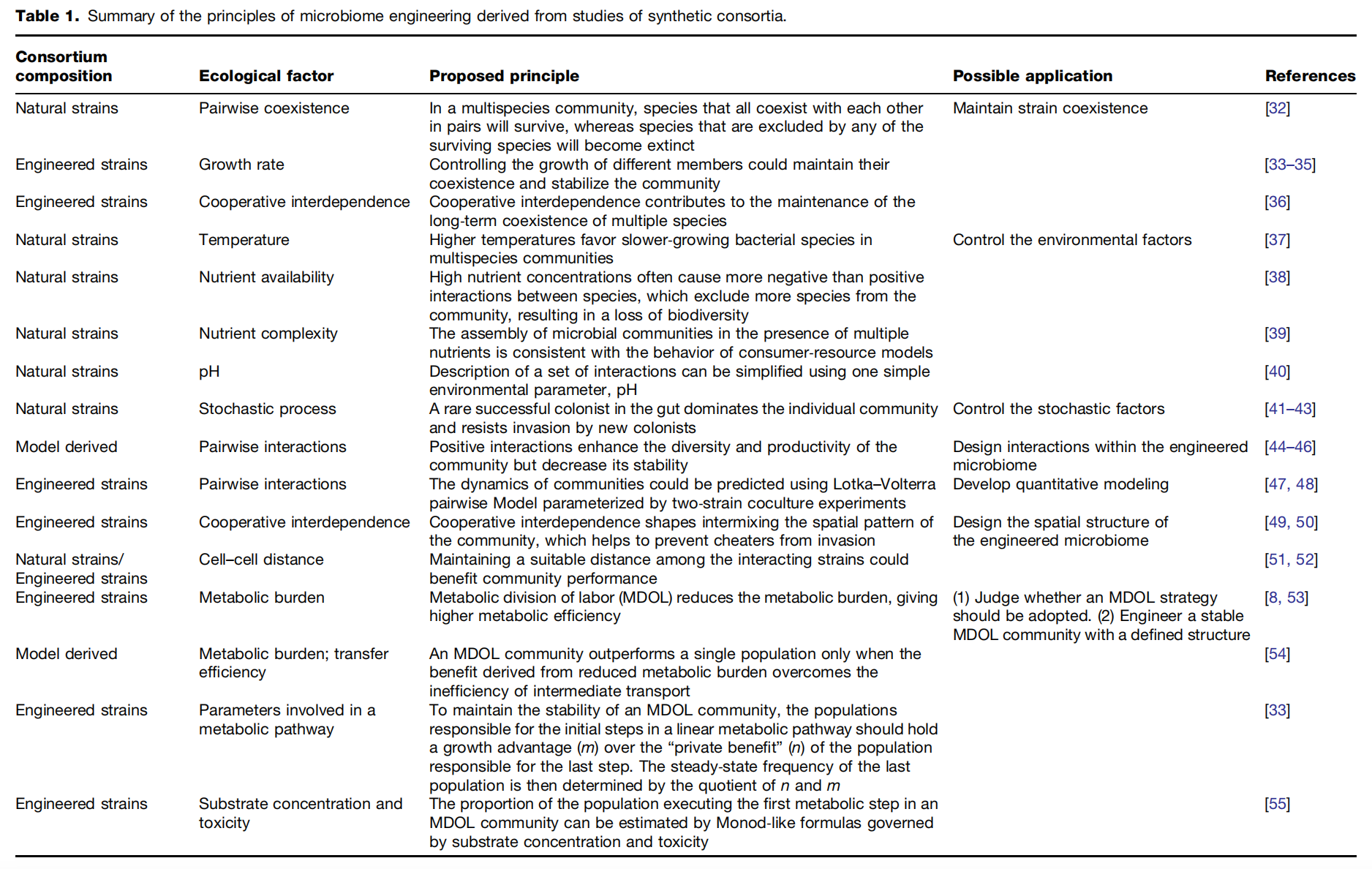

Figure 2 Three methods, enrichment, artificial selection, and directed evolution, are used for top-down microbiome engineering. These methods are designed to drive the self-assembly of a seeding microbiome into an engineered microbiome with the desired functions. (A) During enrichment, the seeding microbiome is introduced into the multiple growth-dilution cycles under well-defined environmental conditions. The microbiome is expected to gradually adapt to the environment and thus develop higher performance101. (B) In artificial selection, a collection of low-density newborn communities is allowed to grow (maturation) into adult communities within a given time period. Then, the adult communities with enhanced functions are chosen to reproduce a novel generation of newborn communities to start the next cycle103. (C) One iteration of directed evolution starts with building a library of generationally stable communities with varied functions, and the community with the highest function is subjected to ecological perturbations110. As a result, a new library of generationally stable communities is generated to start a new iteration. As shown in the right panel of every graph, all these methods can be conceptualized as the shifts of a dynamic structure–function landscape, which represents the changes in the states with specific community structures and functions. Enrichment drives the evolution of the seeding microbiome along one trajectory into a final state with a generationally stable structure and an expected high function. The artificial selection first generates multiple states, and then, the states with higher functions are artificially chosen to continue the selection. As a result, a state with the highest function evolves from diverse evolving trajectories. In addition to the self-assembled states, directed evolution also generates states by imposing ecological perturbations on the self-assembled states, leading to more trajectories that benefit the selection of best-performing communities.
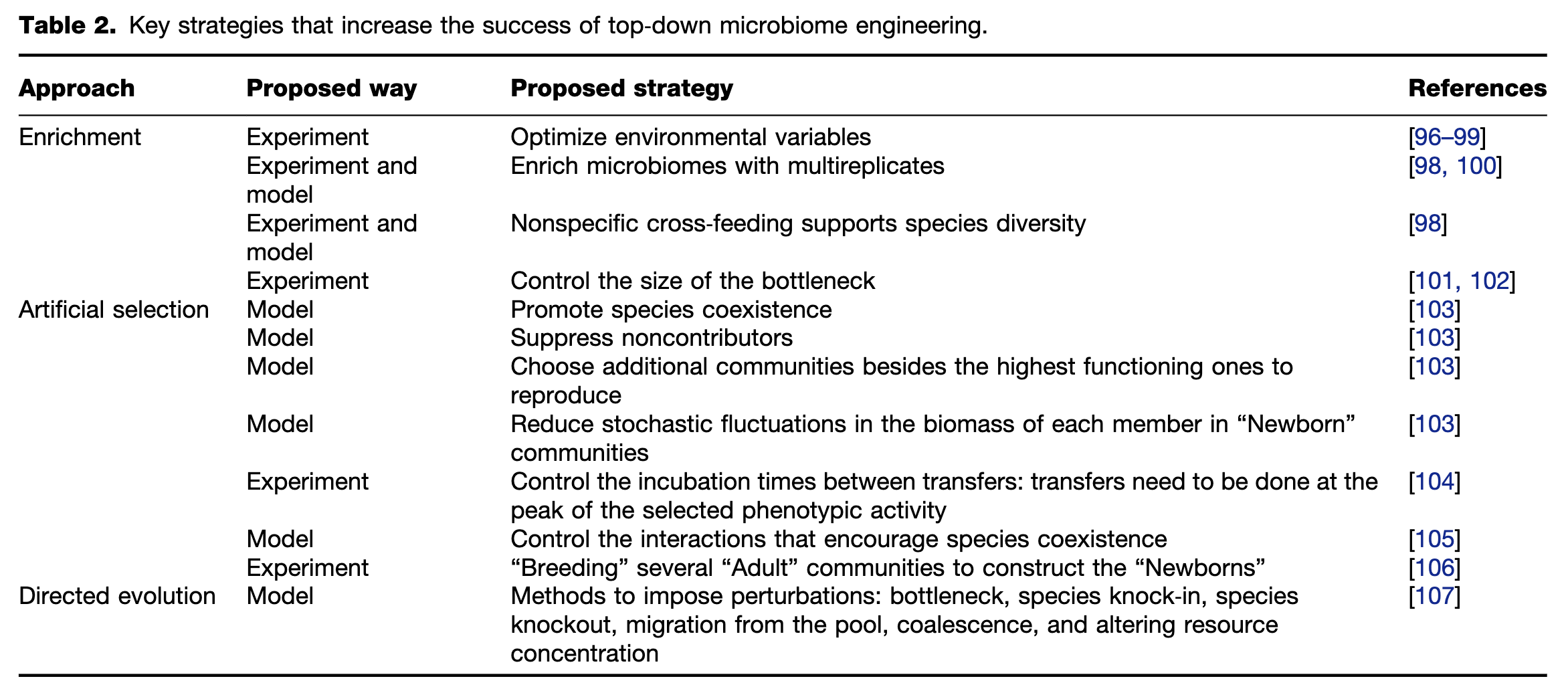

Figure 3 Schematic diagram for the combined framework of top-down and bottom-up approaches for environmental use. (i) Preliminary microbial community with a degrading capability of interest is derived from a seeding microbiome by one or several top-down DBTL cycle(s), in which enrichment, artificial selection, or directed evolution could be applied. (ii) Mechanistic insights of this top-down engineered microbiome can be identified by meta-omics (metagenomics, metatranscriptomics, metaproteomics, or metabolomics), which is incorporated into mathematical modeling to predict interactions among the members. (iii) Based on the mechanistic insights, microorganisms of interest can be isolated by enrichment or high-throughput single-cell sorting and then used for further bottom-up microbiome engineering. (iv) Bottom-up design-build-test-learn (DBTL) cycle(s) would be conducted, guided by mechanistic insights, to engineer a new bottom-up microbiome with higher degrading efficiency and assessed in its effectiveness in the intended application.
https://wap.sciencenet.cn/blog-2675068-1426102.html
上一篇:1979 BioScience 阐明生态系统中的相互作用
下一篇:2023 ER 新型酯酶的表征以及构建能够降解双(2-羟乙基)对苯二甲酸酯的红球菌-伯克霍尔德氏菌菌群